The PopData type
For the PopGen.jl package to be consistent, a standard flexible data structure needs to be defined. The solution is a custom type called PopData. The struct is defined in PopGenCore.jl as:
struct PopData
metadata::PopDataInfo
genodata::DataFrame
end
where PopDataInfo is a nested object defined in PopGenCore.jl as:
mutable struct PopDataInfo
samples::Int64
sampeinfo::DataFrame
loci::Int64
locusinfo::DataFrame
populations::Int64
ploidy::Union{Int8, Vector{Int8}}
biallelic::Bool
end
As you can see, a PopData is made up of two components, one called metadata for sample and locus information, and the other called genodata which includes genotype information. This structure allows for easy and convenient access to the fields using dot . accessors. Both metadata and genodata are specific in their structure, so here is an illustration to help you visualize a PopData object:
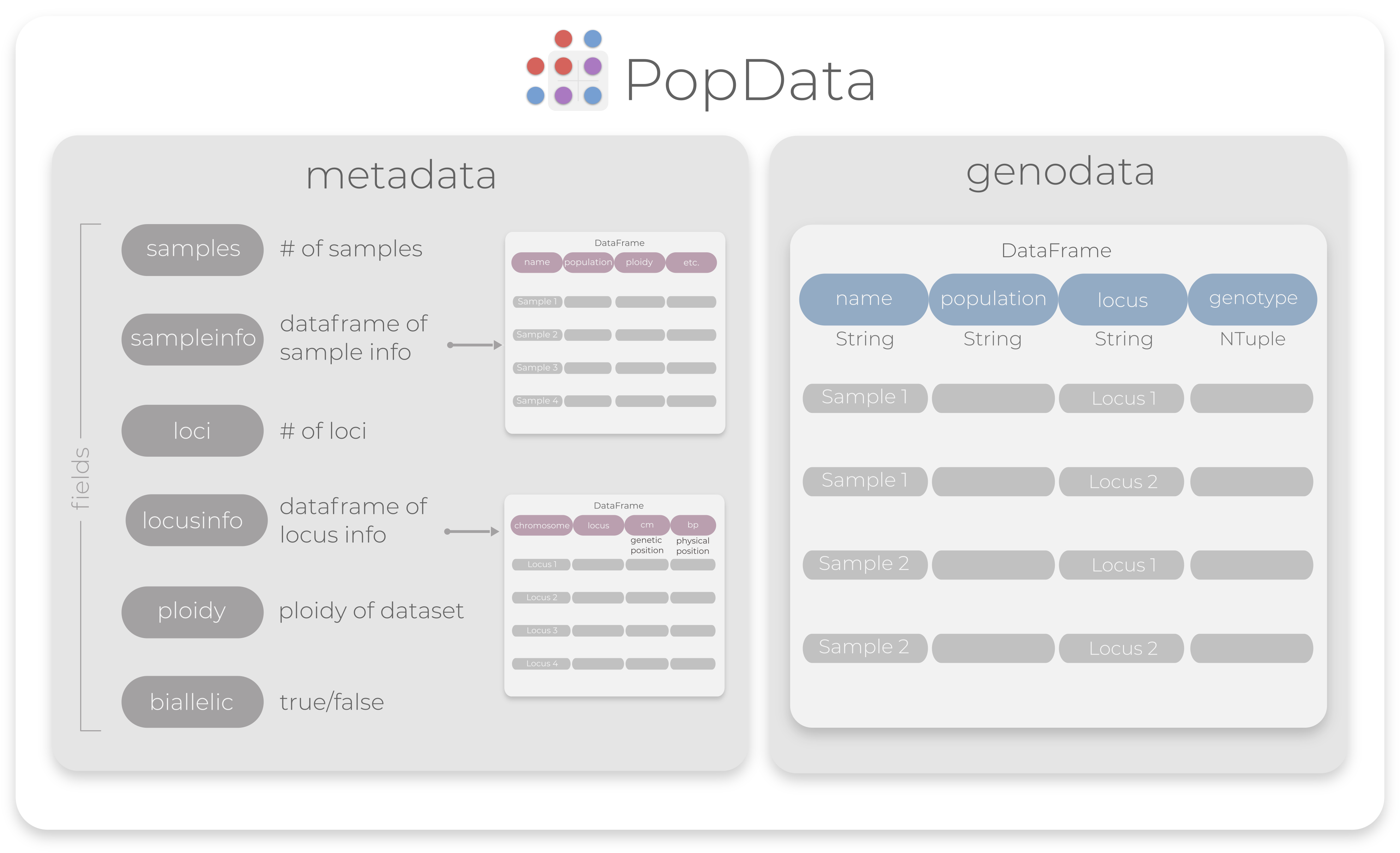
PopData falls under an AbstractType we call PopObj, which is short for "PopGen Object". While not implemented yet,
PopObj exists to futureproof some flexibility into niche data types. Fun fact: we decided to pronounce PopObj as "pop ob" with a silent j because it sounds better than saying "pop obj", but writing it as PopOb looks weird. It's a silly little detail that Pavel seems to care a lot about.
Metadata
The metadata component exists to
- frontload commonly used values like the number of populations and if the data is biallelic
- store information about samples or loci that may be relevant in specific applications
- provide flexibility in storing that information in wide format because storing it in long format would dramatically increase the size of
PopDataobjects - provide easy access to viewing sample or locus information
See viewing data for a closer look at accessing this information.
Genodata
The genotype information is stored in a separate table lovingly called genodata. This table is rather special in that it is stored in "long" format, i.e. one record per row. Storing data this way makes it a lot easier to interrogate the data and write new functions. It also means the table will have as many rows as loci x samples, which can become a lot. To reduce redundant objects inflating object size, the columns name, population, and locus are each a special type of compressed vector from PooledArrays.jl, which is a memory-saving data structure for long repetitive categorical data. Without using this format, gulfsharks, whose source file is 3.2mb, would occupy about 27mb in your RAM! The classes of .genodata can be directly accessed with PopData.genodata.colname where PopData is the name of your PopData object, and colname is one of name, population, locus, genotype. See Advanced Indexing for a deeper
dive into manipulating genodata.
We use the Tuple type for genotypes of individuals because they are immutable (cannot be changed). By the time you're using PopGen.jl, your data should already be filtered and screened. Hand-editing of genotype alleles is strongly discouraged, so we outlawed it.
Other Data Types
While not strictly their own composite types, we also define aliases for genotypes and vectors of genotypes, as their explicit types can get a little unwieldy to use. The types shown below in the code blocks include their name and type (all types are of type DataType) on the first line, and what the alias actually defines on the second line.
Genotype
Genotype::DataType
NTuple{N,<:Signed} where N
An NTuple is itself an alias for a Tuple{Vararg{}} , but you can think of it as Tuple of N length composed of items of a particular type, in this case it's items that are subtypes of Signed (the integer types). The length of the tuple (N) will vary based on the ploidy of the sample, and the element Type will vary whether the markers are snps (Int8) or microsatellites (Int16), making this a pretty flexible (but immutable) structure.
SNP and Msat
Snp::NTuple{N,Int8} where N
Msat::NTuple{N,Int16} where N
These are convenience aliases for the two main kinds of NTuples of genotypes you will see. These are typically used internally.
GenoArray
GenoArray::DataType
AbstractVector{S} where S<:Union{Missing,Genotype}
As you can guess from the name, this Type wraps Genotype into a Vector, while permitting missing values (what's genetics without missing data!?). By using AbstractVector (rather than Vector), we also have the flexibility of functions working on things like SubArrays out of the box.
Getting the most out of Julia and demonstrating good practices means making sure functions work on the things they're supposed to, and give informative error messages when the input isn't suitable for the function (a rare case of wanting MethodErrors). Without these aliases, functions would either have vague definitions like f(x,y,z) where x <: AbstractArray and potentially cause errors, or overly complicated definitions like f(x::AbstractVector{S},y,z) where {N, T<:Signed,S<:NTuple{N,T}} and not be very legible. Instead, functions are written as f(x,y,z) where x<:GenotypeArray, and that seems like a good compromise of getting the latter while looking like the former.
Acknowledgements
A lot of what's possible in PopGen.jl is thanks to the tireless work of the contributors and maintainers of DataFrames.jl. It's no small task to come up with and maintain a robust, performant, and sensible tabular data type, and they deserve so much credit for it.