Comparison
There's a reason we started investing so many hours and so many new grey hairs into writing PopGen.jl when there was an existing ecosystem in R to perform these same tasks. Like we explain in the home page of these docs, we want a platform that is:
- fast(er)
- written in a single language
- easy to use
So, we'd like to prove that Julia and PopGen.jl actually achieves that by showing a few benchmarks comparing PopGen.jl to popular population genetics packages in R. It's worth mentioning that we ourselves use and have published work incorporating these packages, and are incredibly grateful for the work invested in them. We appreciate those folks and have tremendous respect and envy for the work they continue to do! Here are links to adegenet, pegas, hierfstat, and ape.
Benchmarks
To make this a practical comparison, we're going to use the gulfsharks data because it is considerably larger (212 samples x 2209 loci) than nancycats (237 x 9) and a bit more of a "stress test". All benchmarks in R are performed using the microbenchmark package, and BenchmarkTools are used for Julia.
- load Julia packages
- load R packages
using BenchmarkTools, PopGen
library(adegenet)
library(pegas)
library(hierfstat)
library(microbenchmark)
As a note, the reported benchmarks are being performed on a 64-bit Manjaro Linux system on a nothing-special Lenovo Thinkbook 14S with 8gigs of RAM and a 8th gen Intel i5 mobile processor. Note: all of the Julia benchmarks, unless explicitly stated, are performed single-threaded (i.e. not parallel, distributed, or GPU).
Reading in data
Since gulfsharks is provided in PopGenCore.jl, we will just load it with genepop(). If you would like to try this yourself in R, find the gulfsharks.gen file in the package repository under /data/source/gulfsharks.gen. Since the file importer now uses CSV.jl to read files, there is one step of the genepop parser that is multithreaded. However, the majority of the data parsing (formatting the raw data into a correct PopData structure) occurs using a single thread. This R benchmark will take a few minutes. Consider making some tea while you wait.
- Julia
- R
@benchmark sharks = genepop("data/source/gulfsharks.gen", silent = true)
BenchmarkTools.Trial: 10 samples with 1 evaluation.
Range (min … max): 472.671 ms … 526.777 ms ┊ GC (min … max): 0.00% … 5.75%
Time (median): 507.187 ms ┊ GC (median): 5.00%
Time (mean ± σ): 501.984 ms ± 17.301 ms ┊ GC (mean ± σ): 3.47% ± 2.99%
▁ ▁ ▁ ▁ ▁ ▁ █ ▁ ▁
█▁▁▁▁▁▁▁▁▁▁█▁▁▁▁▁█▁▁▁█▁▁▁▁▁▁▁▁▁▁▁▁█▁▁▁▁▁▁▁▁█▁█▁▁▁█▁▁▁▁▁▁▁▁▁▁█ ▁
473 ms Histogram: frequency by time 527 ms <
Memory estimate: 172.46 MiB, allocs estimate: 2246870.
> microbenchmark(read.genepop(file = "/home/pdimens/gulfsharks.gen", ncode = 3L, quiet = TRUE))
Unit: seconds
read.genepop(file = "/home/pdimens/gulfsharks.gen", ncode = 3L, quiet = FALSE)
min lq mean median uq max neval
5.670637 6.218719 6.745065 6.387936 7.019667 9.173005 100
microbenchmark)
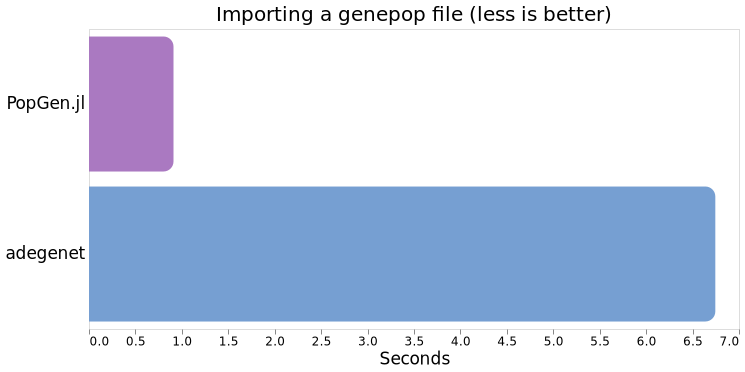
Comparing averages, Julia (via PopGen.jl) clocks in at 507ms versus R (via adegenet) at 6.745s , so ~13.3x faster.
PopData vs genind size
It was pretty tricky to come up with a sensible/efficient/convenient data structure for PopGen.jl, and while the two-DataFrames design might not seem like it took a lot of effort, we ultimately decided that the column-major style and available tools, combined with careful genotype Typing was a decent "middle-ground" of ease-of-use vs performance.
Anyway, it's important to understand how much space your data will take up in memory (your RAM) when you load it in, especially since data's only getting bigger! Keep in mind that gulfsharks in PopGen.jl also provides lat/long data, which should inflate the size of the object somewhat compared to the genind, which we won't add any location data to.
- Julia
- R
julia> Base.summarysize(sharks)
3452870
#bytes
> object.size(gen)
5331536 bytes
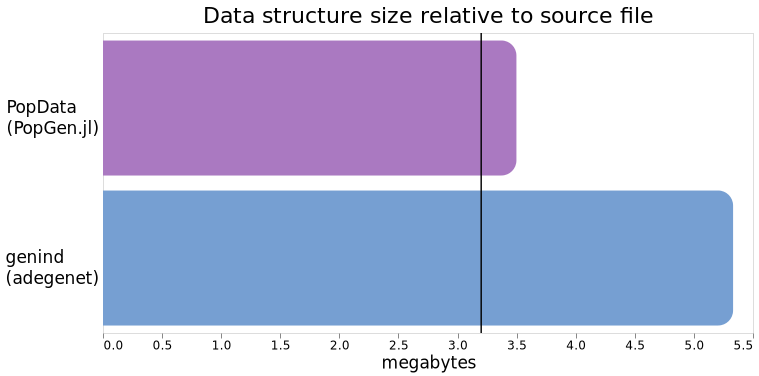
The original genepop file is 3.2mb (the vertical line), and our PopData object takes up ~3.45mb in memory (300kb larger than the source file) versus the ~5.3mb of a genind, which is ~1.5x larger than the source file. That's quite a big difference!
Summary statistics
The obvious hallmark of population genetics is heterozygosity values and F-statistics. Here we'll compare the basic summary statistics that can be produced using hierfstat and PopGen.jl.
- Julia
- R
julia> @benchmark summary(sharks, by = "global")
BenchmarkTools.Trial:
memory estimate: 88.42 MiB
allocs estimate: 1307128
--------------
minimum time: 151.963 ms (0.00% GC)
median time: 171.484 ms (7.60% GC)
mean time: 172.456 ms (6.08% GC)
maximum time: 186.606 ms (7.04% GC)
--------------
samples: 29
evals/sample: 1
> microbenchmark(basic.stats(gen))
Unit: seconds
expr min lq mean
basic.stats(gen) 4.276996 4.425934 4.618796
median uq max neval
4.609901 4.706666 5.292831 100
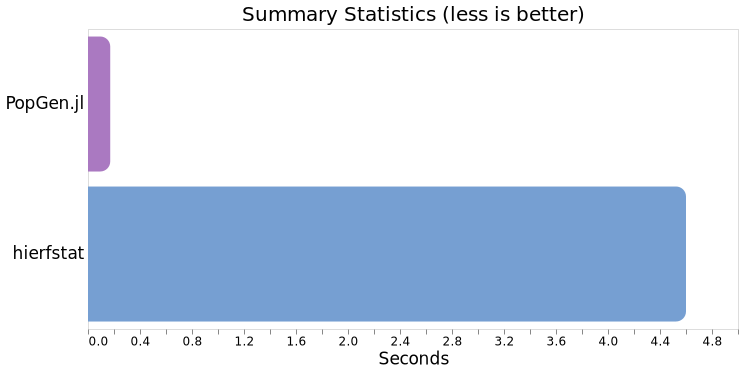
Comparing averages, PopGen.jl clocks in at ~171ms versus hierfstat's 4.6s, which is ~27x faster on these data. However, when testing on a data that was 401 samples x 5331 loci (not shown), PopGen.jl performed 36.6x faster. This gap seems to increase the larger the data is, but we have not tested the upper limits of this.
Chi-squared test for HWE
This is a classic population genetics test and a relatively simple one. The R benchmark will take a while again, so if you're following along, this would be a good time to reconnect with an old friend.
- Julia
- R
julia> @benchmark hwe_test(sharks)
BenchmarkTools.Trial:
memory estimate: 46.22 MiB
allocs estimate: 1050525
--------------
minimum time: 145.476 ms (0.00% GC)
median time: 179.146 ms (4.35% GC)
mean time: 176.142 ms (3.56% GC)
maximum time: 204.813 ms (0.00% GC)
--------------
samples: 29
evals/sample: 1
> microbenchmark(hw.test(gen, B = 0))
Unit: seconds
expr min lq mean median uq max neval
hw.test(gen, B = 0) 5.100298 5.564807 6.265948 5.878842 6.917006 8.815179 100
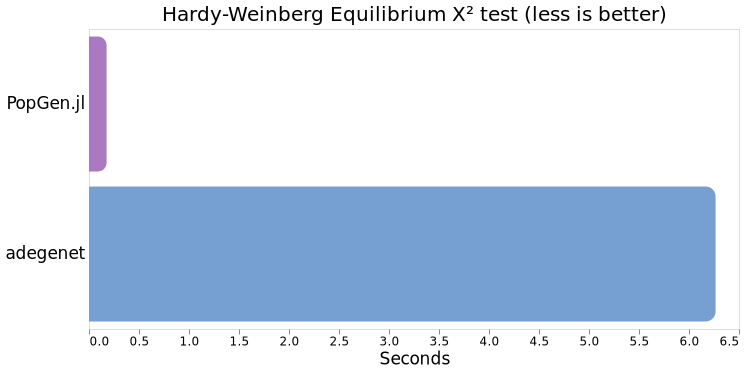
Comparing averages, PopGen.jl clocks in at ~176ms versus adegenet's 6.3s, so ~35.8x faster on these data(!)
Pairwise FST
You all know it, you all love it. What's population genetics without a little pairwise FST sprinkled in? This benchmark compairs the Weir & Cockerham pairwise FST calculation in PopGen.jl against hierfstat
- Julia
- Julia (parallel)
- R
We will add the extra keywords samples and seconds to the benchmark
macro so we can get a full 100 evaluations. You will need to start Julia with 1 available threads via julia --threads 1 (julia >= v1.5) or JULIA_NUM_THREADS=1 (< v1.5).
julia> @benchmark pairwise_fst(sharks) samples = 100 seconds = 700
BenchmarkTools.Trial:
memory estimate: 869.93 MiB
allocs estimate: 6090633
--------------
minimum time: 557.995 ms (9.29% GC)
median time: 571.297 ms (11.26% GC)
mean time: 580.627 ms (12.59% GC)
maximum time: 754.451 ms (31.41% GC)
--------------
samples: 100
evals/sample: 1
This is to demonstrate what the speed is like when starting Julia with 6 available threads via julia --threads 6 (julia >= v1.5) or JULIA_NUM_THREADS=6 (< v1.5).
julia> @benchmark pairwise_fst(sharks) samples = 100 seconds = 700
BenchmarkTools.Trial:
memory estimate: 869.93 MiB
allocs estimate: 6090639
--------------
minimum time: 205.038 ms (0.00% GC)
median time: 305.189 ms (28.90% GC)
mean time: 299.227 ms (25.03% GC)
maximum time: 359.663 ms (27.05% GC)
--------------
samples: 100
evals/sample: 1
We'll need to convert sharks into the matrix/dataframe hierfstat needs
to run this calculation. The conversion will be a separate step so as not
to add unnecessary (or unfair) overhead to the benchmark. This benchmark is
going to take forever (200s/run x 100 runs = 5.5hrs), so if you absolutely insist on
trying it out yourself, you may want to pop outside and enjoy some fresh
air for a bit (I ran it overnight). Seriously, you don't want to watch this paint dry 🖌️.
> sharks_hierf <- genind2hierfstat(sharks)
> microbenchmark(pairwise.WCfst(sharks_hierf))
Unit: seconds
expr min lq mean median uq max neval
pairwise.WCfst(sharks2) 192.2786 192.9277 199.4861 193.5743 195.0079 301.6879 100
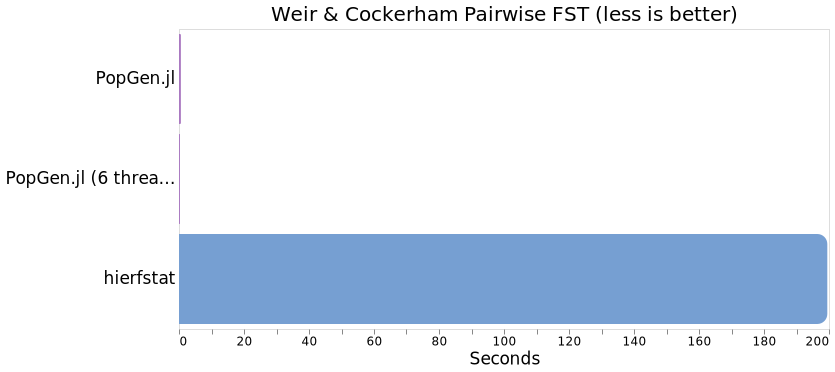
On a single thread, pairwise FST in PopGen.jl is ~340x faster than in hierfstat, and a whopping 665x faster using 6 threads with the optimized matrix-based implementation.